Joanna Niska-Blakie, PhD
Global Product Manager
Qiagen
Poland
Introduction
Manufacturing therapeutics, especially viral vectors for cell and gene therapy, demands rigorous safety standards and strict regulatory compliance. Key challenges include efficient cell lysis and the thorough removal of impurities such as host cell DNA (hcDNA), which can compromise product quality and safety. Regulatory agencies such as the FDA, WHO and EMA enforce strict limits on residual DNA in final products to manage these risks. High-salt environments help reduce viscosity during purification, enhancing process efficiency. Salt-active endonuclease Saltonase® GMP-grade is an effective tool that leverages these conditions to degrade nucleic acids efficiently. Combined with Saltonase ELISA Kit (5 x 96), these tools ensure robust purification workflows, improved product quality and process-related impurity control in adherence to regulatory requirements.
Cell lysis and its role in viral vector purification
In the early stages of viral vector purification, eukaryotic cells are lysed using a buffer formulated with pH stabilizers, mild detergents and high-salt concentrations. The elevated salt not only enhances cell lysis but also minimizes vector aggregation and lowers solution viscosity, all of which contribute to more efficient downstream processing.
Following lysis, the mixture contains viral particles alongside both target and genomic DNA. To reduce contamination and facilitate subsequent purification steps, salt-active endonucleases optimize the digestion of the hcDNA impurities (see Figure 1).
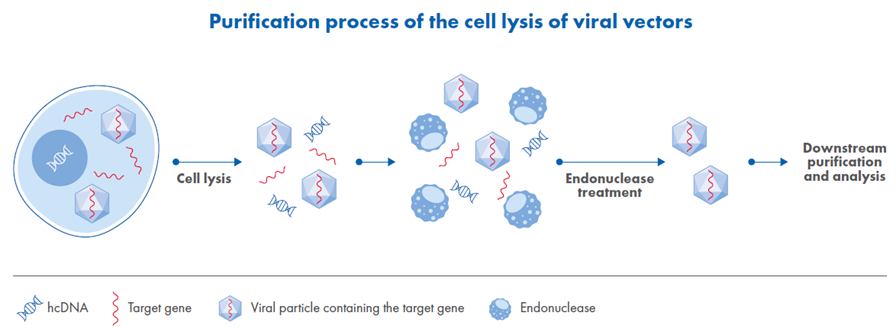
Figure 1. Overview of viral vector purification beginning with high-salt cell lysis.
Risks Associated with Residual Host Cell DNA
Even low levels of residual DNA can compromise product safety and reduce manufacturing efficiency, making its removal a critical step in gene therapy production. Residual hcDNA in biopharmaceutical products poses serious safety and manufacturing challenges. One primary concern is oncogenicity; if leftover DNA fragments contain oncogenes, they could potentially integrate into a patient’s genome and trigger tumor formation. Random DNA integration can disrupt essential regulatory genes even without oncogenes, increasing the risk of harmful mutations.
There is also a virological risk, as residual DNA might lead to the creation of infectious particles if viral genomes integrate incorrectly or unwanted sequences propagate. From a manufacturing standpoint, free DNA increases solution viscosity, which makes purification steps like filtration and chromatography more difficult. This can cause membrane clogging, slow down processing, raise material costs and result in product loss.
To manage these risks, regulatory agencies such as the FDA, WHO and EMA enforce strict limits on residual DNA in final products. Most biological products listed in the European Pharmacopoeia are subject to a general residual DNA limit of no more than 10 ng per dose. However, certain vaccines have stricter requirements – for instance, the residual DNA content in inactivated hepatitis A vaccine must not exceed 100 pg per dose, while for the hepatitis B vaccine, the limit is even lower at 10 pg per dose. These limits reflect the lower biological activity and integration risk of shorter DNA fragments.1
Because of these stringent requirements, manufacturers take a cautious approach, removing as much residual DNA as possible to ensure safety, meet regulatory standards and maintain efficient production.
Challenges in DNA Degradation During Lysis
Removing DNA during cell lysis is critical but presents several challenges. One key obstacle is the tightly packed structure of DNA within chromatin. DNA is wrapped around histone proteins, creating a compact formation that limits enzyme access. Even after lysis, these histone-DNA interactions often remain, reducing the effectiveness of conventional endonucleases.
Another challenge is the high viscosity caused by large DNA molecules in the solution. This increased viscosity interferes with mixing and processing and slows enzyme diffusion, which lowers their efficiency. However, using high-salt concentrations in the lysis buffer can help. Salt promotes chromatin decondensation and reduces viscosity, improving enzyme access to DNA and ultimately boosting viral vector yields
Cost is also an important consideration. Endonucleases are relatively expensive, so maximizing their efficiency is essential for keeping the process economical. Enzymes with high specific activity allow for lower doses without compromising performance, helping to reduce overall manufacturing costs.
Saltonase GMP-Grade: A Novel Solution for DNA Removal
To address these challenges, QIAGEN developed Saltonase GMP-grade, a next-generation, salt-active endonuclease specifically optimized for high-salt lysis environments. Unlike conventional enzymes, Saltonase was engineered from a psychrophilic bacterial source and is expressed in Escherichia coli, offering both scalability and reliability.
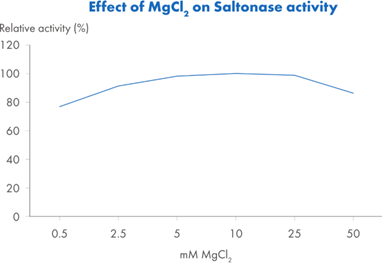
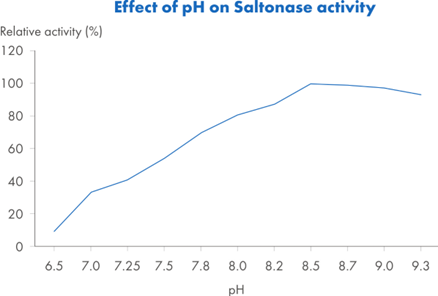
Saltonase distinguishes itself with a molecular weight of 40 kDa and the ability to maintain activity across a broad range of bioprocessing conditions. It performs optimally in buffers containing 500 mM NaCl, at a pH of 8.5 and a temperature of 37°C, with at least 1 mM MgCl2 present. However, it retains at least 20% relative activity across a wide operational window (Figure 2): salt concentrations from 0.1 to 0.9 M, temperatures from 15°C to 55°C, and pH values ranging from 6.8 to 9.3. This versatility makes it well-suited for integration into various upstream workflows without requiring extensive buffer adjustments or additional processing steps.
| A. |
B. |
 |
 |
| C. |
D. |
 |
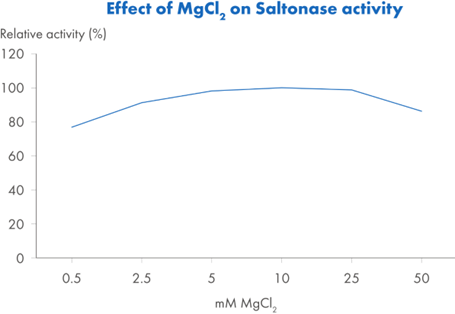 |
Figure 2. Relative activity* of Saltonase across: Figure 2A. Salt concentrations 0–900 mM NaCl, with constant pH 8.5, T=37°C and MgCl2 5mM. Figure 2B. pH: 6.5–9.3, with constant NaCl 500 mM, T=37°C and MgCl2 5mM. Figure 2C. Temperature 4–80°C, with constant NaCl 500 mM, pH 8.5 and MgCl2 5 mM. Figure 2D. MgCl2 concentrations 0.5–50 mM, with constant NaCl 500 mM, pH 8.5, and T=37°C.
*Relative activity is a ratio of a test sample vs Saltonase at its optimal activity measured at NaCl 500 mM, pH 8.5, T=37 °C and MgCl2 5mM.
Digestion Robustness of Saltonase Makes it The Safest Endonuclease Choice
To assess the robustness and efficiency of Saltonase, we performed a comparative digestion assay of Saltonase and a widely used salt-intolerant endonuclease. The assay used dsDNA substrates tagged with fluorophores and quenchers to monitor DNA cleavage in real time. When the endonuclease cuts the DNA, the quencher is separated from the fluorophore, allowing fluorescence to be emitted. This increase in signal, measured in relative fluorescence units (RFUs), directly reflects the extent of DNA digestion.
The experiment included both 10 bp and 15 bp probes to evaluate the degree of fragmentation. Higher RFU values indicate more efficient DNA breakdown. As shown in Figure 3, Saltonase demonstrated superior performance, cleaving DNA into fragments as short as 3–5 nucleotides, highlighting its effectiveness in applications where thorough DNA removal is essential.

Figure 3. The digestion efficiency of Saltonase and a salt-intolerant endonuclease from Supplier M was assessed using a dsDNA substrate labeled with a quencher in close proximity to a fluorophore. Subsequent cleavage of the substrate resulted in a fluorescence signal. The experiments were conducted at a temperature of 23°C, chosen due to the low melting temperature (Tm) of the probes. Each reaction utilized 0.4 U of enzyme. Measurements were taken at 495 nm excitation and 525 nm emission, with a 515 nm cut-off for fluorescence detection. RFU – Relative Fluorescence Units.
Performance in Variable Salt and pH Environments
Saltonase was tested alongside two other endonucleases, one salt-tolerant and one salt-intolerant, across a range of NaCl concentrations (0, 150 and 500 mM) and pH values (6.8, 7.4 and 8.5), at both 37 °C and room temperature.
As shown in Figure 4, Saltonase consistently outperformed both enzymes under all tested conditions. Its activity remained high even at elevated salt levels, particularly at physiological and alkaline pH. Notably, under more acidic conditions, Saltonase showed superior performance compared to the other enzymes at NaCl concentrations above 300 mM. These results highlight its versatility and effectiveness across diverse bioprocessing environments.
Saltonase Applicability
Saltonase has a broad applicability in viral vector purification, including adeno-associated virus (AAV), lentivirus (LV) and virus-like particles (VLPs). In the quality control of recombinant AAV (rAAV) vectors, essential for ensuring safety and efficacy in gene therapy, Saltonase plays a key role in reducing residual DNA contaminants, which are common byproducts of vector production.
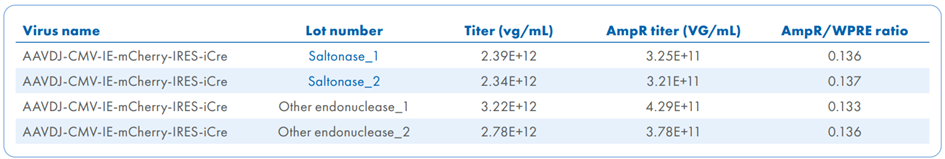
In this study (Figure 5), AAV samples were distributed across four plates and treated with Saltonase and other salt-active endonucleases. Each batch was analyzed for DNA contamination using dPCR (Figure 5 A), while capsid protein profiles were assessed through protein staining (Figure 5 B). The standard deviation of the AmpR/WPRE ratio was lower in Saltonase-treated samples (SD = 0.0007) compared to those treated with the alternative salt-active endonuclease (SD = 0.002), indicating greater consistency in plasmid DNA removal. Functional validation was conducted through cell-based transduction assays, followed by DAPI nuclear staining to assess transgene expression and cellular localization and dsRed to confirm successful transduction (Figure 5 C). This integrated approach confirmed the effectiveness of Saltonase in supporting high-quality rAAV production.
Figure 4. Activity (U/µL) of Saltonase, salt-tolerant endonuclease and salt-intolerant endonuclease conducted at three NaCl concentrations (0, 150, 500 mM) across three pH levels (6.8, 7.4, 8.5) at 37 °C and 23 °C.
A.
B.
C.
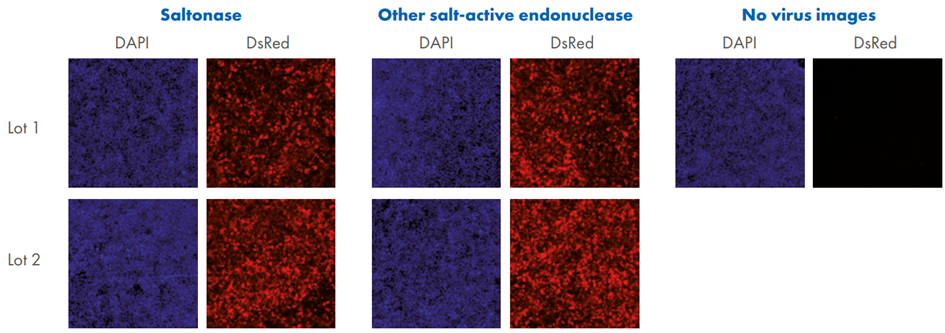
Figure 5. Quantification and quality assessment of AAV produced using different endonucleases. A. Four individual 1-plate AAV packaging runs were performed using two different salt-active endonucleases: Saltonase and another salt-active endonuclease, with two AAV lots produced per enzyme. Each lot was evaluated by dPCR using primer/probe sets targeting WPRE (viral genome) and AmpR (plasmid DNA). B. For protein quality control, an aliquot from each AAV lot was analyzed by SDS-PAGE followed by SimplyBlue Safe Stain using a microwave protocol. Gels were imaged under 685 nm wavelength on an Azure Sapphire system to assess the presence and integrity of AAV capsid proteins. C. HEK293 cells were seeded at 5 × 104 cells/well in a 96-well plate (200 µL/well). Approximately 22 hours later, 100 µL of media was removed, and 5 µL of AAV was added to each well (three wells per AAV lot). Two hours post-transduction, 100 µL of fresh media was replenished. At 24 hours post-transduction, cells were fixed and stained. Fluorescent imaging was performed using an Evident APEX microscope. NV – no virus (AAV) added to control cells stained with DAPI and dsRed.
LV and VLP Purification Workflows
Saltonase has demonstrated exceptional performance under a range of challenging bioprocessing conditions, underscoring its versatility and robustness. In lentiviral vector purification workflows, Saltonase retained high enzymatic activity at a concentration of 50 U/mL in the presence of 150 mM NaCl, at pH 7.0, and at a low temperature of 8°C over a 12–hour incubation period, conditions often considered suboptimal for enzymatic digestion. Despite these constraints, it achieved an impressive 99.8% reduction in hcDNA (Figure 6 A).
Similarly, in virus-like particle (VLP) processing, Saltonase delivered rapid and effective DNA degradation, reaching over 86% removal within just 45 minutes at room temperature, under elevated salt conditions (500 mM NaCl) and slightly acidic pH (6.5) (Figure 6 B). These results highlight Saltonase’s adaptability and consistent performance across diverse and demanding production environments, making it a reliable tool for nucleic acid removal in advanced biomanufacturing workflows.
A.
B.
Figure 6. Efficiency of Saltonase in host cell DNA removal in (A) lentiviral purification and (B) virus-like particle purification.
Detecting Residual Enzyme Levels
Effective control of process-related impurities is a critical step in biomanufacturing to ensure the safety and quality of therapeutic products. After its use in bioprocessing, Saltonase must be thoroughly removed through standard downstream purification techniques such as depth filtration, diafiltration and chromatography. These steps not only eliminate cellular debris and contaminants but also target residual enzymatic additives like Saltonase. Given the stringent sensitivity required to detect trace amounts of residual enzyme, robust analytical methods are essential to verify complete removal. To address this need, QIAGEN has developed the Saltonase ELISA Kit (5 x 96), a highly sensitive and specific assay that employs monoclonal antibodies with strong affinity for Saltonase. This kit enables detection of residual enzyme at levels as low as 30 pg/mL, making it ideal for routine in-process and final product testing. By ensuring precise quantification of residual Saltonase, the ELISA kit supports regulatory compliance and helps minimize immunogenic risks, underscoring its vital role in impurity control during biomanufacturing.
Conclusion
Efficient removal of hcDNA and process-related enzymes is essential for safe, compliant, and high-yield viral vector manufacturing. By combining salt-active enzymatic digestion with sensitive detection tools, biomanufacturers can achieve robust impurity control even under challenging conditions.
Together, Saltonase GMP-grade and the Saltonase ELISA Kit provide a powerful, end-to-end solution for optimizing purification workflows and meeting regulatory standards with confidence.
References
1 World Health Organization. (2005, April). Report of the WHO informal consultation on molecular methods to support the quality of vaccines and biological products (WHO/BS/05.1994). https://www.who.int/docs/default-source/biologicals/vaccine-quality/69-molecular-methods-final-mtg-report-april2005.pdf
#IndustryMemberNews